CellR4 2014; 2 (2): e856
Personalized Medicine and Surgery
Topic: Cell Differentiation
Category: Reviews
Abstract
MATERIALS AND METHODS: Expert opinion supplemented by literature search.
RESULTS: The infrastructure of a successful personalized medicine center should have four essential components: 1) genomic/molecular data acquisition and storage, 2) integration of genomic diagnostic testing and targeted imaging, 3) research focused on functional genomic targets, and 4) development and informed use of targeted therapies of actionable genes.
CONCLUSIONS: There is strong academic and public interest in advancing personalized medicine, which promises more precise, efficient patient care. Moving forward, clear and centralized consensus on actionable genes is needed.
Introduction
Modern advances in DNA sequencing, tissue banking, and electronic medical records are moving personalized medicine to the forefront of healthcare. Personalized medicine refers to the tailoring of medical and surgical therapies to the individual’s unique molecular profile. This represents a more precise, focused approach compared to current clinical medicine, which is based upon studies collected from large, heterogeneous populations of patients. Just as CT scanning has revolutionized medicine by providing greater detail about a patient’s internal organs, genetic profiling now provides greater detail of molecular-level anatomy allowing clinicians to better diagnose preclinical disease and predict response to therapy. In current practice, molecularly diverse patients are often grouped and assigned the same treatment, knowing a minority of patients will receive no benefit or will be exposed to adverse effects of therapy. Customization of therapy to an individual’s genome and particular set of enzymes will improve efficacy and reduce the number of treatment failures. Personalized medicine promises to vastly improve patient care by customizing current treatments, diagnosing disease earlier, and facilitating development and implementation of targeted therapies.
Governmental, academic, and specialty organizations have endorsed the development of personalized medicine. The US Department of Health and Human Services (HHS) Secretary’s Priorities states, “HHS supports research on personalized medicine… [and] more effective individualized prevention efforts, as well as methods to deliver the right care to the right patient at the right time” 1. To this end, HHS has funded efforts by the CDC and NIH to establish the 100 most significant genes pertaining to public health, as well as a number of genome-wide disease association studies 2. In 2008, the Secretary’s Advisory Committee on Genetics, Health, and Society highlighted the importance of pharmacogenomics, a subtype of personalized medicine 3. Consequently, academic medical centers have been rapidly creating and expanding centers of personalized medicine, have held a number of personalized medicine conferences, and have focused on personalized medicine and surgery at national surgical conferences 4 , 5 , 6 , 7 , 8. National multidisciplinary organizations, most notably the Personalized Medicine Coalition, also champion personalized medicine. Overall, there is a strong desire amongst governmental and academic institutions, as well as the general public, to fully realize the potential of personalized medicine.
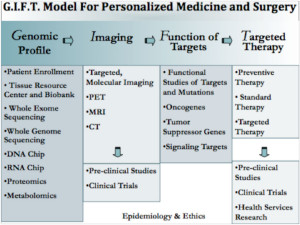
This article reviews the elements needed to establish a successful personalized medicine program. These elements are based on our model for establishing a center for personalized medicine and surgery, the GIFT model, which details the process in four discrete phases 9 (see Figure 1). Phase 1, genomic profiling, is establishing an infrastructure to collect and store patients’ molecular profile, which includes DNA, RNA, proteomic, and epigenetic data. Phase 2, imaging, describes the immediate integration and application of patient molecular profile into clinical decision-making. Phase 3, function of targets, genomic data are used in novel research techniques to identify previously unknown disease-associated molecular derangements. Phase 4, targeted therapy, utilizes the information gained from functional analysis towards the development of new targeted therapies, which include small molecule, immunologic, surgical, and/or gene therapies based on the individual patient’s personalized molecular profile.
Genomic Data/Molecular Profile
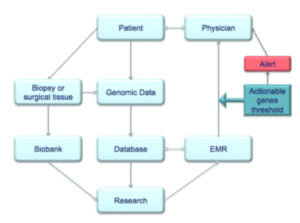
Personalized medicine tailors care to the patient’s molecular profile. Thus, the ability to collect, store, analyze, and efficiently utilize patients’ genomic information is paramount (see Figure 2). Centers of personalized medicine must integrate DNA sequencing, molecular profiling, electronic medical records, and biobanking.
The modern driving force behind personalized medicine is the ability to rapidly sequence DNA. Underpinning all of today’s sequencing techniques is the work done by the Human Genome Project (HGP). Started in 1984 and formally funded in 1990, the HGP was a collaboration by the National Institute of Health and the Department of Energy to completely sequence all 3 million base pairs of the human genome. Not only did the HGP succeed in sequencing the human genome, it inspired significant advancements in rapid DNA sequencing. One of the first patients to have their entire genome sequenced was the co-discoverer of DNA structure, Dr. James Watson. His genome was sequenced in 4 months at a cost of $1.5 million dollars 10. In comparison, the human genome project took 13 years and $6 billion dollars. Growth of sequencing capability has progressed, and currently the human genome can be sequenced in days or weeks for less than $1,000 dollars 11.
Repeated genomic sequence testing may be necessary in an individual patient to detect changes over time, or to test for different diagnoses. Thus, rapid, cost efficient sequencing is vital. To accomplish this, several large academic centers have established DNA sequencing core facilities to expedite sequencing and minimize costs. Some of these centers also incorporate innovative high throughput DNA sequencing methods, such as pyrosequencing and the Genome Analyzer system (Illumina, San Diego, CA, USA). These techniques provide increased processing speed at the expense of an increased error rate 12. Ideally, the commercialization of DNA sequencing will allow centers without associated sequencing core labs to access this technology to drive down costs. Another up and coming method of is to use DNA microarrays, also known as DNA “chips”. Microarrays sequence pre-selected genes related to a target disease by DNA-probe hybridization and quantification 13 , 14. The focused approach reduces cost but requires pre-selected target sequences, providing less information. Cost effective, universal whole genome sequencing is the ideal all centers should strive for.
Evaluation of certain diseases will require gathering information beyond genomic data. DNA is only a portion of the larger molecular profile, which contains proteomic, RNA, and epigenetic information. Non-DNA molecular information is clinically relevant; for example, testing for the Her2/neu protein has become routine in breast cancer care. Her2/neu positivity determines whether patients should receive Her2/neu targeted therapies such as trastuzumab. Her2 amplification is caused by over-expression, not mutation or deletion, of the ERBB2 gene. Thus, this effect would go undetected by DNA sequencing. RNA expression is another important aspect of the molecular profile. For example, serum microRNA are currently being studied as a marker of colonic adenomas 15 and CEACAM-7 mRNA has been proposed as a potential marker of rectal cancer recurrence. Moreover, a number of diagnostic assays for Lupus, Thyroid Cancer, hypertension, and chronic lymphocytic leukemia (CLL) based upon mRNA expression levels have been described 16 , 17 , 18 , 19. Finally, epigenetic modifications are involved in Albright’s hereditary osteodystrophy, Beckwith-Widemann, Prader-Willi, and Angelman syndromes. Studies of the inflammatory bowel diseases have demonstrated the genetic component is responsible for only 8-13% of disease variance, and believe it may be attributed to epigenetic changes 20. Increased DNA methylation in inflammatory bowel disease due to high cell turnover also contributes to the predisposition to develop cancer 21. For many diseases, future centers of personalized medicine will require evaluation of the entire molecular profile.
Currently, our ability to gather genomic data has outpaced our ability to interpret it. Thus, transforming the deluge of information into meaningful clinical data requires integration with electronic medical records (EMRs). Several pilot programs merging personalized patient data to EMRs have been established. These programs highlight the key components of EMR integration: 1) availability of genomic testing, 2) robust electronic medical record, 3) pre-determined actionable genes set by multidisciplinary consensus panels based upon current literature, and 4) patient privacy safeguards. CLIPMERGE PGx is an innovative program based at Mount Sinai Medical Center that integrates the center’s institutional biobank with its electronic medical record; pre-determined “actionable” patient genotypes automatically generate electronic alerts with advice for the patient’s provider 22. Vanderbilt, the University of Florida, and Stanford have also established similar programs 23 , 24. Two elements of EMR integration, genomic testing and a robust EMR, are widely available at large medical centers and are expanding to smaller health facilities. A significant obstacle is that current actionable gene lists are built in a decentralized and institution-specific fashion. Moving forward, the NIH-CDC Scientific Foundation for Personal Genomics recommends that an independent group such as the United States Preventative Services Task Force (USPSTF) or Evaluation of Genomic Applications in Practice and Prevention (EGAPP) should create a universal actionable gene list 25.
Biobanks are another key component of a personalized medicine center, which serve a dual role patient care and research. Many medical centers have already developed robust biobanks 26 , 27 , 28 , 29 , 30 and the NIH has considered establishing a national biobank 31. First, centralized storage of patient blood and tissue samples allow for future testing or auto-transplantation. Thus, collaboration with pathology to ensure proper handling and storage of tissues would be optimal. Second, they provide researchers access to a wide assortment of tissues for molecular profiling to investigate derangements associated with disease states. In order to avoid labor-intensive consent procedures for every use of the stored tissue, biobanks commonly obtain blanket informed consent for any number of future research studies to be performed with the collected tissue or generated data. While this is a huge boon to researchers and clinicians, patients are vulnerable to violations of privacy and care must be taken to keep patients informed of relevant findings from various studies.
It is important to consider patient privacy and right-to-information during the development of a personalized medicine program. Privacy concerns should be addressed pre-emptively. Patient genomic information should be carefully communicated to patients, and information given to family members and outside facilities should be done with great sensitivity. A communication infrastructure within the EMR to notify patients about important results or research efforts should be established. Patients should also be reassured that the enactment of the Genetic Information Nondiscrimination Act (GINA) in 2008 prevents employers or health insurance providers from discriminating against individuals based upon their genetic information.
Diagnosis, Prognostication, and Imaging
The current immediate clinical applications of personalized genomic data are to aid diagnosis, determine drug response via pharmacogenomics, and perform novel imaging techniques. Focusing on these modalities will help bridge personalized medicine to clinical practice.
Diagnostic implications are vast for disease-defining genes. Prime candidates for genome wide screening would include elective newborn screening, patients with suspected disease associated with disease-defining genetic derangements (particularly oncologic diseases), and patients undergoing expensive or critical medical therapies where drug metabolism will impact outcomes. Genomic analysis, mainly in the form of karyotyping and now chromosomal microarray, is already a primary diagnostic tool for obstetricians 32. Expectant parents are already offered expanded, non-invasive, prenatal genetic screening for various genetic diseases; this may soon replace invasive testing and be expanded to whole genome sequencing 33. In adult patients, there are many indications for genetic testing of specific genes in a multitude of cancers, as well as many non-oncologic hereditary disorders such as Brugada syndrome, hereditary angioedema, and hemochromatosis. Additionally, genomic testing can serve as diagnostic aids in areas of uncertainty. For example, many fine-needle aspirations performed for a thyroid nodule that are categorized as “atypia of undetermined significance” or “follicular lesion of unknown significance” have a 15-30% chance of being malignant, leading to a large number of unnecessary thyroidectomies. The Veracyte test is a panel of 167 genes with reported negative predictive value of 94-95% 34, which promises to help prevent many nontherapeutic thyroid surgeries. Veracyte also illustrates how genomic testing can also reduce or replace more expensive or invasive tests or procedures. Successful implementation also requires that providers be properly educated on the uses of genomic information. Inappropriate use of biomarkers, such as the widespread use of prostate specific antigen as a screening tool, can lead to unnecessary procedures, morbidity, and costs 35.
Prognostication of disease and predicting drug responses are also important applications of personalized medicine. Oncotype DX and MammaPrint are existing gene-expression profiling assays designed to estimate disease recurrence and potential benefit of adjuvant therapy. A similar genetic screen designed to predict disease recurrence in patients with high-risk prostate cancer has been proposed 36. Predicting the course of a malignancy can help guide important decision-making processes in cancer care, such as determining the aggressiveness of surgical therapy and the value of adjuvant therapies. Genetics can also be used to predict the potential benefit of pharmacologic therapy or customize medications and endpoints of therapy for high-risk individuals. Characterization of an individual’s profile of cytochrome p450 enzymes affects metabolism of clopidogrel, warfarin, simvastatin, and many other medications. As many as 16-50% of patients prescribed clopidogrel are either non-responsive or minimally responsive to the medication, which in turn is associated with increased cardiovascular thrombotic events, particularly after coronary stenting 37. Furthermore, the BENEDICT study found that variants in the gene ADAMST13 predict endovascular complications due to diabetes and the percentage of patients that benefit from an ACE inhibitor 38. These examples emphasize how using genetic data to predict response to drugs could significantly impact health outcomes.
Molecular imaging exploits variations in the molecular profile and combines them with current imaging technologies. This type of functional imaging is exemplified in 18F-FDG PET scanning, radioactive iodine imaging of thyroid disease, or imaging of pheochromocytomas with 18F-L-dopa, which utilizes the unique uptake of labeled glucose, iodine, or tyrosine precursors, respectively. Development of further imaging techniques follows the same concept: labeling molecules that are differentially metabolized in certain disease states. Tumor-specific or tumor-associated cell markers, such as CA 19-9, can be targeted with labeled antibodies for PET imaging 39. Tyrosine kinases are overactive in a number of cancers, including melanoma, lung, and leukemia 40. By targeting this specific alteration, 11C-erlotinib PET scanning can identify residual increased tyrosine kinase activity, signifying refractory disease and predicting to tyrosine kinase-inhibitor therapy 41.
Functional genomic research
Previously, for any disease process, suspected genes of interest were selected and tested one by one. Genome sequencing now allows comparison of genomes of patients with and without disease to reveal associated mutations, providing a genomic map of target sequences to investigate. This has lead to several breakthroughs in poorly understood diseases, such as dopamine-sensitive dystonia. Genome-wide comparison identified an unexpected mutation in calcium channel gene CACNA1S found to be the cause of dystonia 42. Now, dozens of genome-wide association studies have been done for a multitude of diseases including asthma, diabetes, and heart disease 43 , 44. Recently whole exome sequencing of 142 pancreatic cancer samples was able to highlight significant alterations in SLIT/ROBO signaling in early stage pancreatic cancers 45. Though these studies may not yet directly translate into clinical practice, they provide a roadmap to guide further investigation of mutated sequences that have a high likelihood of giving rise to disease.
Evaluation of differential gene expression via mRNA signals is another important tool in genomic research. This is routinely done with RNA microarrays, which are chips with short sequences complementary to target mRNA sequences. These are able to report which mRNA are significantly elevated or decreased in pathologic tissue samples. Unlike genome sequencing, target RNA sequences need to be preselected and incorporated into the RNA microarray chip. Genes that are over-expressed can be targeted with RNA interference or small molecules to interrupt their associated biochemical pathways. However, since gene expression is influenced by many ever-changing factors such as hypoxia and serum glucose levels, there is significant noise in microarray data. Furthermore, since mRNA can vary greatly between tissues types, including of a variety of cell types in a single tissue sample increases noise as well. Using laser dissection techniques to carve out specific cells for testing can potentially reduce this source of error, but also makes RNA isolation more difficult. Identifying meaningful differentially expressed genes requires a large number of samples to avoid false results, emphasizing the utility of personalized medicine center with a large repository of data.
Both aforementioned processes generate a large amount of data, most of which remains clinically insignificant and requires sophisticated interpretation. Filtering of data from these experiments is greatly enhanced by software such as DAVID (Database for Annotation, Visualization, and Integrated Discovery), which is a collection of free, online tools that help sort lists of genes or proteins into known biochemical pathways. DAVID relies on other online databases such as the Kyoto Encyclopedia of Genes and Genomes, which is a large, updated collection of molecular pathways. Thus, a strong systems biology arm is vital in interpreting gene lists from the aforementioned studies. These “top-down” novel investigative protocols help researchers hone in on genes in known relevant pathways, identify novel potentially oncologic pathways, and facilitate more focused and traditional research protocols.
Therapies for Actionable Genomics
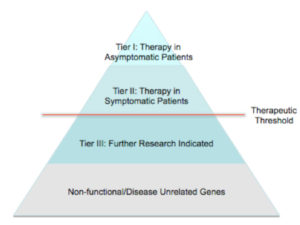
The ultimate goal of personalized medicine is tailored medical and surgical therapy based on a patient’s molecular profile. When a particular genetic mutation or variation in molecular profile rises above certain thresholds for medical or surgical intervention, it should dictate therapy. Genes that warrant clinical intervention we have termed “actionable genomics.” We believe actionable genes should be separated into tiers, including 1) genes that warrant action in an asymptomatic patient, 2) genes that warrant action in patients with symptomatic disease, and 3) genes related to disease, but requiring further investigation (Figure 3).
The first tier, actionable genes in asymptomatic patients, encompasses many established examples in various cancer oncogenes and lethal congenital conditions. Activating RET mutations now dictates early thyroidectomy for medullary thyroid cancer; patients with E-cadherin mutations should undergo prophylactic gastrectomy; BRCA1 and BRCA2 mutations make it reasonable to pursue prophylactic mastectomy. Other well-known examples include prophylactic management of phenylketonuria (PKU) and cystic fibrosis. These genes confer either a high or definite likelihood of disease with well-tolerated medical or surgical treatments.
For second tier genes, treatments are reserved until patients have begun to manifest symptoms of disease. These include cancers with aberrancies in molecular profile eligible for targeted therapy. Some examples include Crizotinib for anaplastic lymphoma kinase (ALK) re-arrangement in non-small cell lung carcinomas (NSCLC) 46 and Ipilimumab and Vemurafenib in BRAF activating mutations 47 , 48. Other diseases in this tier have a disease defining gene mutation, but the treatment is morbid and reserved for late stages of disease. This includes alpha-1 antitrypsin deficiency, Wilson disease, familial amyloidosis, and glycogen storage diseases that can cause end stage liver failure requiring liver transplantation for select cases 49.
Third tier genes are simply those that are currently under investigation. Many third tier genes are likely derived from genome-wide association studies or RNA microarray research, where they are frequently altered in disease but their role in disease or function is still unclear. These simply require verification through traditional genomic research techniques.
Conclusions
There is strong academic and public interest in advancing personalized medicine, which promises more precise, efficient patient care. The infrastructure of a successful personalized medicine center should have four essential components: 1) genomic/molecular data acquisition and storage, 2) integration of genomic diagnostic testing and targeted imaging, 3) research focused on functional genomic targets, and 4) development and informed use of targeted therapies of actionable genes. Moving forward, clear and centralized consensus on actionable genes is needed.
Conflict of Interests: The Authors declare that they have no conflict of interests.
References
- Services, U.S.D.o.H.H. Accelerate the Process of Scientific Discovery To Improve Patient Care. [cited 2013 11/17]; Available from: http://www.hhs.gov/secretary/about/priorities/accelerate.html (back)
- Office, H.P. HHS Secretary Leavitt Announces Steps Toward A Future of “Personalized Health Care”. March 23, 2007 [cited 2013 11/17]; Available from: http://www.hhs.gov/news/press/2007pres/20070323a.html (back)
- Secretary’s Advisory Committee on Genetics, H., and Society, Realizing the Potential of Pharmacogenomics: Opportunities and Challenges. May 2008. (back)
- Farrugia G, Weinshilboum RM. Challenges in implementing genomic medicine: the Mayo Clinic Center for Individualized Medicine. Clin Pharmacol Ther 2013; 94(2): 204-206. (back)
- Brunicardi FC, Gibbs RA, Fisher W, Chen C. Overview of the Molecular Surgeon Symposium on Personalized Genomic Medicine and Surgery. World J Surg 2009; 33(4): 612-614. (back)
- Meric-Bernstam F, Farhangfar C, Mendelsohn J, Mills GB. Building a personalized medicine infrastructure at a major cancer center. J Clin Oncol 2013; 31(15): 1849-1857. (back)
- Fenstermacher DA, Wenham RM, Rollison DE, Dalton WS. Implementing personalized medicine in a cancer center. Cancer J 2011; 17(6): 528-536. (back)
- Brunicardi FC. Molecular surgery and biology. Am J Surg 2000; 180(6): 397-401. (back)
- Brunicardi FC, Gibbs RA, Fisher W, Chen C. Overview of the Molecular Surgeon Symposium on Personalized Genomic Medicine and Surgery. World J Surg 2009; 33(4): 612-614. (back)
- Wadman M. James Watson’s genome sequenced at high speed. Nature 2008; 452(7189): 788. (back)
- Johnson JA, Burkley BM, Langaee TY, Clare-Salzler MJ, Klein TE, Altman RB. Implementing personalized medicine: development of a cost-effective customized pharmacogenetics genotyping array. Clin Pharmacol Ther 2012; 92(4): 437-439. (back)
- Voidonikolas G, Kreml SS, Chen C, Fisher WE, Brunicardi FC, Gibbs RA, Gingras MC. Basic principles and technologies for deciphering the genetic map of cancer. World J Surg 2009; 33(4): 615-629. (back)
- Selman M, Pardo A, Barrera L, Estrada A, Watson SR, Wilson K, Aziz N, Kaminski N, Zlotnik A. Gene expression profiles distinguish idiopathic pulmonary fibrosis from hypersensitivity pneumonitis. Am J Respir Crit Care Med 2006; 173(2): 188-198. (back)
- Radmacher MD, Marcucci G, Ruppert AS, Mrózek K, Whitman SP, Vardiman JW, Paschka P, Vukosavljevic T, Baldus CD, Kolitz JE, Caligiuri MA, Larson RA, Bloomfield CD; Cancer and Leukemia Group B. Independent confirmation of a prognostic gene-expression signature in adult acute myeloid leukemia with a normal karyotype: a Cancer and Leukemia Group B study. Blood 2006; 108(5): 1677-1683. (back)
- Kanaan Z, Roberts H, Eichenberger MR, Billeter A, Ocheretner G, Pan J, Rai SN, Jorden J, Williford A, Galandiuk S. A plasma microRNA panel for detection of colorectal adenomas: a step toward more precise screening for colorectal cancer. Ann Surg 2013; 258(3): 400-408. (back)
- Carlsen AL, Schetter AJ, Nielsen CT, Lood C, Knudsen S, Voss A, Harris CC, Hellmark T, Segelmark M, Jacobsen S, Bengtsson AA, Heegaard NH. Circulating microRNA expression profiles associated with systemic lupus erythematosus. Arthritis Rheum 2013; 65(5): 1324-1334. (back)
- Jacques C, Guillotin D, Fontaine JF, Franc B, Mirebeau-Prunier D, Fleury A, Malthiery Y, Savagner F. DNA microarray and miRNA analyses reinforce the classification of follicular thyroid tumors. J Clin Endocrinol Metab 2013; 98(5): E981-989. (back)
- Visone R, Rassenti LZ, Veronese A, Taccioli C, Costinean S, Aguda BD, Volinia S, Ferracin M, Palatini J, Balatti V, Alder H, Negrini M, Kipps TJ, Croce CM. Karyotype-specific microRNA signature in chronic lymphocytic leukemia. Blood 2009; 114(18): 3872-3879. (back)
- Li S, Zhu J, Zhang W, Chen Y, Zhang K, Popescu LM, Ma X, Lau WB, Rong R, Yu X, Wang B, Li Y, Xiao C, Zhang M, Wang S, Yu L, Chen AF, Yang X, Cai J. Signature microRNA expression profile of essential hypertension and its novel link to human cytomegalovirus infection. Circulation 2011; 124(2): 175-184. (back)
- Ventham NT, Kennedy NA, Nimmo ER, Satsangi J. Beyond gene discovery in inflammatory bowel disease: the emerging role of epigenetics. Gastroenterology 2013; 145(2): 293-308. (back)
- Ventham NT, Kennedy NA, Nimmo ER, Satsangi J. Beyond gene discovery in inflammatory bowel disease: the emerging role of epigenetics. Gastroenterology 2013; 145(2): 293-308. (back)
- Gottesman O, Scott SA, Ellis SB, Overby CL, Ludtke A, Hulot JS, Hall J, Chatani K, Myers K, Kannry JL, Bottinger EP. The CLIPMERGE PGx Program: clinical implementation of personalized medicine through electronic health records and genomics-pharmacogenomics. Clin Pharmacol Ther 2013; 94(2): 214-217. (back)
- Johnson JA, Burkley BM, Langaee TY, Clare-Salzler MJ, Klein TE, Altman RB. Implementing personalized medicine: development of a cost-effective customized pharmacogenetics genotyping array. Clin Pharmacol Ther 2012; 92(4): 437-439. (back)
- Pulley JM, Denny JC, Peterson JF, Bernard GR, Vnencak-Jones CL, Ramirez AH, Delaney JT, Bowton E, Brothers K, Johnson K, Crawford DC, Schildcrout J, Masys DR, Dilks HH, Wilke RA, Clayton EW, Shultz E, Laposata M, McPherson J, Jirjis JN, Roden DM. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin Pharmacol Ther 2012; 92(1): 87-95. (back)
- Khoury MJ, McBride CM, Schully SD, Ioannidis JP, Feero WG, Janssens AC, Gwinn M, Simons-Morton DG, Bernhardt JM, Cargill M, Chanock SJ, Church GM, Coates RJ, Collins FS, Croyle RT, Davis BR, Downing GJ, Duross A, Friedman S, Gail MH, Ginsburg GS, Green RC, Greene MH, Greenland P, Gulcher JR, Hsu A, Hudson KL, Kardia SL, Kimmel PL, Lauer MS, Miller AM, Offit K, Ransohoff DF, Roberts JS, Rasooly RS, Stefansson K, Terry SF, Teutsch SM, Trepanier A, Wanke KL, Witte JS, Xu J; Centers for Disease Control and Prevention. The Scientific Foundation for personal genomics: recommendations from a National Institutes of Health-Centers for Disease Control and Prevention multidisciplinary workshop. Genet Med 2009; 11(8): 559-567. (back)
- Olson JE, Ryu E, Johnson KJ, Koenig BA, Maschke KJ, Morrisette JA, Liebow M, Takahashi PY, Fredericksen ZS, Sharma RG, Anderson KS, Hathcock MA, Carnahan JA, Pathak J, Lindor NM, Beebe TJ, Thibodeau SN, Cerhan JR. The Mayo Clinic Biobank: a building block for individualized medicine. Mayo Clin Proc 2013; 88(9): 952-962. (back)
- Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, Masys DR. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther 2008; 84(3): 362-369. (back)
- Harring TR, Guiteau JJ, Nguyen NT, Cotton RT, Gingras MC, Wheeler DA, O’Mahony CA, Gibbs RA, Brunicardi FC, Goss JA. Building a comprehensive genomic program for hepatocellular carcinoma. World J Surg 2011; 35(8): 1746-1750. (back)
- Nguyen NT, Cotton RT, Harring TR, Guiteau JJ, Gingras MC, Wheeler DA, O’Mahony CA, Gibbs RA, Brunicardi FC, Goss JA. A primer on a hepatocellular carcinoma bioresource bank using the cancer genome atlas guidelines: practical issues and pitfalls. World J Surg 2011; 35(8): 1732-1737. (back)
- Voidonikolas G, Gingras MC, Hodges S, McGuire AL, Chen C, Gibbs RA, Brunicardi FC, Fisher WE. Developing a tissue resource to characterize the genome of pancreatic cancer. World J Surg 2009; 33(4): 723-731. (back)
- Murphy J, Scott J, Kaufman D, Geller G, LeRoy L, Hudson K. Public perspectives on informed consent for biobanking. Am J Public Health 2009; 99(12): 2128-2134. (back)
- Wapner RJ, Martin CL, Levy B, Ballif BC, Eng CM, Zachary JM, Savage M, Platt LD, Saltzman D, Grobman WA, Klugman S, Scholl T, Simpson JL, McCall K, Aggarwal VS, Bunke B, Nahum O, Patel A, Lamb AN, Thom EA, Beaudet AL, Ledbetter DH, Shaffer LG, Jackson L. Chromosomal microarray versus karyotyping for prenatal diagnosis. N Engl J Med 2012; 367(23): 2175-2184. (back)
- Benn P, Chapman AR, Erickson K, Defrancesco MS, Wilkins-Haug L, Egan JF, Schulkin J. Obstetricians’ and gynecologists’ practice and opinions of expanded carrier testing and non-invasive prenatal testing. Prenat Diagn 2013 Nov 13. doi: 10.1002/pd.4272. [Epub ahead of print] (back)
- Alexander EK, Kennedy GC, Baloch ZW, Cibas ES, Chudova D, Diggans J, Friedman L, Kloos RT, LiVolsi VA, Mandel SJ, Raab SS, Rosai J, Steward DL, Walsh PS, Wilde JI, Zeiger MA, Lanman RB, Haugen BR. Preoperative diagnosis of benign thyroid nodules with indeterminate cytology. N Engl J Med 2012; 367(8): 705-715. (back)
- Kerfoot BP, Holmberg EF, Lawler EV, Krupat E, Conlin PR. Practitioner-level determinants of inappropriate prostate-specific antigen screening. Arch Intern Med 2007; 167(13): 1367-1372. (back)
- Karnes RJ, Bergstralh EJ, Davicioni E, Ghadessi M, Buerki C, Mitra AP, Crisan A, Erho N, Vergara IA, Lam LL, Carlson R, Thompson DJ, Haddad Z, Zimmermann B, Sierocinski T, Triche TJ, Kollmeyer T, Ballman KV, Black PC, Klee GG, Jenkins RB. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk patient population. J Urol 2013; 190(6): 2047-2053. (back)
- Udaya Tantry CH, Zehnder J, Gurbel P. Clopidogrel resistance and clopidogrel treatment failure. 2013; Available from: http://www.uptodate.com/contents/clopidogrel-resistance-and-clopidogrel-treatment-failure (back)
- Rossing P, Lajer M. Can ADAMTS13 lead us to the paradise of personalized medicine? Diabetes 2013; 62(10): 3331-3332. (back)
- Girgis MD, Federman N, Rochefort MM, McCabe KE, Wu AM, Nagy JO, Denny C, Tomlinson JS. An engineered anti-CA19-9 cys-diabody for positron emission tomography imaging of pancreatic cancer and targeting of polymerized liposomal nanoparticles. J Surg Res 2013; 185(1): 45-55. (back)
- Poot AJ, Slobbe P, Hendrikse NH, Windhorst AD, van Dongen GA. Imaging of TKI-target interactions for personalized cancer therapy. Clin Pharmacol Ther 2013; 93(3): 239-241. (back)
- Meng X, Loo BW Jr, Ma L, Murphy JD, Sun X, Yu J. Molecular imaging with 11C-PD153035 PET/CT predicts survival in non-small cell lung cancer treated with EGFR-TKI: a pilot study. J Nucl Med 2011; 52(10): 1573-1579. (back)
- Hanchard NA, Murdock DR, Magoulas PL, Bainbridge M, Muzny D, Wu YQ, Wang M, McGuire AL, Lupski JR, Gibbs RA, Brown CW. Exploring the utility of whole-exome sequencing as a diagnostic tool in a child with atypical episodic muscle weakness. Clin Genet 2013; 83(5): 457-461. (back)
- Ng MC, Saxena R, Li J, Palmer ND, Dimitrov L, Xu J, Rasmussen-Torvik LJ, Zmuda JM, Siscovick DS, Patel SR, Crook ED, Sims M, Chen YD, Bertoni AG, Li M, Grant SF, Dupuis J, Meigs JB, Psaty BM, Pankow JS, Langefeld CD, Freedman BI, Rotter JI, Wilson JG, Bowden DW. Transferability and fine mapping of type 2 diabetes loci in African Americans: the Candidate Gene Association Resource Plus Study. Diabetes, 2013; 62(3): 965-976. (back)
- Li X, Hawkins GA, Ampleford EJ, Moore WC, Li H, Hastie AT, Howard TD, Boushey HA, Busse WW, Calhoun WJ, Castro M, Erzurum SC, Israel E, Lemanske RF Jr, Szefler SJ, Wasserman SI, Wenzel SE, Peters SP, Meyers DA, Bleecker ER. Genome-wide association study identifies TH1 pathway genes associated with lung function in asthmatic patients. J Allergy Clin Immunol 2013; 132(2): 313-20 e15. (back)
- Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J, Chang DK, Cowley MJ, Gardiner BB, Song S, Harliwong I, Idrisoglu S, Nourse C, Nourbakhsh E, Manning S, Wani S, Gongora M, Pajic M, Scarlett CJ, Gill AJ, Pinho AV, Rooman I, Anderson M, Holmes O, Leonard C, Taylor D, Wood S, Xu Q, Nones K, Fink JL, Christ A, Bruxner T, Cloonan N, Kolle G, Newell F, Pinese M, Mead RS, Humphris JL, Kaplan W, Jones MD, Colvin EK, Nagrial AM, Humphrey ES, Chou A, Chin VT, Chantrill LA, Mawson A, Samra JS, Kench JG, Lovell JA, Daly RJ, Merrett ND, Toon C, Epari K, Nguyen NQ, Barbour A, Zeps N; Australian Pancreatic Cancer Genome Initiative, Kakkar N, Zhao F, Wu YQ, Wang M, Muzny DM, Fisher WE, Brunicardi FC, Hodges SE, Reid JG, Drummond J, Chang K, Han Y, Lewis LR, Dinh H, Buhay CJ, Beck T, Timms L, Sam M, Begley K, Brown A, Pai D, Panchal A, Buchner N, De Borja R, Denroche RE, Yung CK, Serra S, Onetto N, Mukhopadhyay D, Tsao MS, Shaw PA, Petersen GM, Gallinger S, Hruban RH, Maitra A, Iacobuzio-Donahue CA, Schulick RD, Wolfgang CL, Morgan RA, Lawlor RT, Capelli P, Corbo V, Scardoni M, Tortora G, Tempero MA, Mann KM, Jenkins NA, Perez-Mancera PA, Adams DJ, Largaespada DA, Wessels LF, Rust AG, Stein LD, Tuveson DA, Copeland NG, Musgrove EA, Scarpa A, Eshleman JR, Hudson TJ, Sutherland RL, Wheeler DA, Pearson JV, McPherson JD, Gibbs RA, Grimmond SM. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012; 491(7424): 399-405. (back)
- Shaw AT, Kim DW, Nakagawa K, Seto T, Crinó L, Ahn MJ, De Pas T, Besse B, Solomon BJ, Blackhall F, Wu YL, Thomas M, O’Byrne KJ, Moro-Sibilot D, Camidge DR, Mok T, Hirsh V, Riely GJ, Iyer S, Tassell V, Polli A, Wilner KD, Jänne PA. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013; 368(25): 2385-2394. (back)
- Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, Akerley W, van den Eertwegh AJ, Lutzky J, Lorigan P, Vaubel JM, Linette GP, Hogg D, Ottensmeier CH, Lebbé C, Peschel C, Quirt I, Clark JI, Wolchok JD, Weber JS, Tian J, Yellin MJ, Nichol GM, Hoos A, Urba WJ. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010; 363(8): 711-723. (back)
- Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O’Dwyer PJ, Lee RJ, Grippo JF, Nolop K, Chapman PB. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med 2010; 363(9): 809-819. (back)
- Petrowsky H, Brunicardi FC, Leow VM, Venick RS, Agopian V, Kaldas FM, Zarrinpar A, Markovic D, McDiarmid SV, Hong JC, Farmer DG, Hiatt JR, Busuttil RW. Liver transplantation for lethal genetic syndromes: a novel model of personalized genomic medicine. J Am Coll Surg 2013; 216(4): 534-543; discussion 543-544. (back)
To cite this article
Personalized Medicine and Surgery
CellR4 2014; 2 (2): e856
Publication History
Published online: 31 Mar 2014

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.